Karine SERRE, PhD
IMMUNOLOGIST
Latest Posts
21/01/2016
Psoriasis is an autoimmune disease and NOT a chronic inflammatory disease!12/05/2015
CALL For PAPERS: “Innate T Cell Development and Functions in Immune Diseases”28/08/2014
XL Annual Meeting of the Sociedade Portuguesa de Imunologia 13-15th October 201413/04/2013
Beginnings of breaking down the differentiation program of innate IL-17-production27/01/2013
The improbable trio NKG2D – γδ T cells – IgE!!CD4 T cell diversification during a single primary immune response
Diversification of the CD4 T helper cell responses during a single immune response
Cohorts of CD4 T helper cells are produced in vivo with individual cells showing functional specialization. This applies to the responses I have studied, where naïve CD4 T cells responding to alum-precipitated protein in lymph nodes give rise to a range of cells producing different cytokines that perform specialized and distinct functions in different sites and at different times after exposure to antigen (1). To study in vivo CD4 T cell responses, transgenic naïve ovalbumin-specific CD4 (OTII) T cells were monitored during their response to alum-precipitated ovalbumin (alumOVA).
Diversification of primed CD4 T cells as assess by cytokine expression
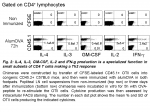
CD4 T helper cells up-regulating cytokines characteristic of Th1 (IFN-g), Th2 (IL-4, IL-13), follicular B helper T cells (IL-21) along with IL-2, IL-3 and GM-CSF secretion are all produced simultaneously in response to alum-precipitated protein (Fig. 1) (2).
In addition, most of these cytokines have been detected at the protein level by intracellular FACS staining after ex vivo restimulation through TCR signalling (Fig. 2).
Therefore, specialisation is apparent in relation with cytokine production as early as 3 days after immunization with alumOVA. The population of CD4 T cells responding to alum-precipitated protein contains Th2-features as well as TFh-features, and to a lesser extent some properties associated with Th1 cells. This system offers the possibility of studying in vivo the emergence of these various helper CD4 T cells. This raises the question as to whether one effector subset produces all these cytokines or whether different effector subsets are generated that produce various combinations of these cytokines. This was addressed in the next section by performing real time RT-PCR at the single cell level on OTII cells primed in vivo.
Substantial diversity within the CD4 Th2 population generated in vivo in response to alum-precipitated protein
Diversity becomes even more apparent when responses are analyzed by measurement of cytokine mRNA expression at the single cell level. This reveals that some aspects are either hidden or overemphasized when the properties are measured at the population level.
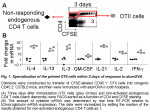
 Thus, I have assessed how Th2-cytokine expressions are orchestrated in Th2 OTII cells primed to alumOVA in vivo. To this end, chimeras were constructed by transfer of naïve CFSE-labeled OTII cells into congenic WT mice. Three days after immunization OTII cells were isolated as single cells at successive stages of differentiation (Fig. 3A). Naïve OTII cells were compared with OTII cells that had upregulated CD69 but had not yet divided, OTII cells that had completed 2-3 divisions and OTII cells that had divided 6 times.
Thus, I have assessed how Th2-cytokine expressions are orchestrated in Th2 OTII cells primed to alumOVA in vivo. To this end, chimeras were constructed by transfer of naïve CFSE-labeled OTII cells into congenic WT mice. Three days after immunization OTII cells were isolated as single cells at successive stages of differentiation (Fig. 3A). Naïve OTII cells were compared with OTII cells that had upregulated CD69 but had not yet divided, OTII cells that had completed 2-3 divisions and OTII cells that had divided 6 times.
Real time RT-PCR was performed on the single cells from these subsets. IL-4 and IL-13 mRNA were manifest only in a small subset ~7% and 2%, respectively, of OTII cells that had undergone 6 divisions (Fig. 3B). This reflects the typical proportion of IL-4-containing OTII cells revealed by intracellular staining in responses to alumOVA (1, 3, 4).
The extensive heterogeneity of cytokine producing cells is further exemplified by the finding that some Th2 cells produce both IL-4 and IL-13, while others produce IL-4 but not IL-13, or IL-13 but not IL-4 (Fig. 3C).
These results exemplify the substantial variety within the CD4 Th2 population, generated in vivo in response to alum-precipitated protein. In this context, I am in the process of extending the number of gene expression analysed by RT-PCR within single cells. Based on a previously published approach (5, 6), I am using this method to measure the expression of several transcription factors, associated with Th2 or TFh development, in relation with the expression of the cytokines described above (IL-2, IL-3, IL-4, IL-13, GM-CSF, and IFN-g). This should give further insight into the selective acquisition of effector functions by CD4 T cells.
Diversification of primed CD4 T cells as assess their localization within the responding LN and through recirculation within the entire body
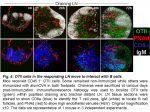 The location of CD4 T cells provides insight into their function. Classically, naïve CD4 T cells occupy the central T zones where they screen the surface of dendritic cells, while primed CD4 T cells in the outer T zone make cognate interaction with B cells that have captured Ag, and follicular CD4 T cells are key to the differentiation and survival of germinal center B cells (7). After transfer into non-immunized mice naïve OTII cells recirculate between the T zones of LN (Fig. 4). By 24h after alumOVA the draining LN has enlarged and the OTII cell numbers are increased. By 48h OTII cell numbers have increased markedly. They are distributed throughout the enlarged T zone and there is now a focus of responding cells in the outer T zone adjacent to the follicles. By 72h many OTII cells have migrated into the follicles and the number in the T zone has further increased. The timing of migration into follicles is in keeping with previous reports (8). The follicular location correlates with strong CXCR5 expression by a proportion of the OTII cells (not shown and (3)).
The location of CD4 T cells provides insight into their function. Classically, naïve CD4 T cells occupy the central T zones where they screen the surface of dendritic cells, while primed CD4 T cells in the outer T zone make cognate interaction with B cells that have captured Ag, and follicular CD4 T cells are key to the differentiation and survival of germinal center B cells (7). After transfer into non-immunized mice naïve OTII cells recirculate between the T zones of LN (Fig. 4). By 24h after alumOVA the draining LN has enlarged and the OTII cell numbers are increased. By 48h OTII cell numbers have increased markedly. They are distributed throughout the enlarged T zone and there is now a focus of responding cells in the outer T zone adjacent to the follicles. By 72h many OTII cells have migrated into the follicles and the number in the T zone has further increased. The timing of migration into follicles is in keeping with previous reports (8). The follicular location correlates with strong CXCR5 expression by a proportion of the OTII cells (not shown and (3)).
 By 72h after immunization, beside B cell follicle localization, some CFSElow OTII cells have left the draining LN and entered both the blood and Ag-free distant LN (Fig. 5). These OTII cells are likely emigrants from the responding LN, for they have completed at least 4 divisions. If they had been activated in the distant LN, some OTII cells that had divided fewer than 4 times would have been observed there from 48h and onwards and these were not present. The primed immigrant OTII cells were present in distant nodes in increased numbers at 96h. These early recirculating OTII cells have features of central memory cells, including expression of CD44 and CD62L. In addition, they re-entered cell cycle and migrated to B cell follicles, much more rapidly than naïve CD4 T cells and could still be induced to produce IL-4 (3).
By 72h after immunization, beside B cell follicle localization, some CFSElow OTII cells have left the draining LN and entered both the blood and Ag-free distant LN (Fig. 5). These OTII cells are likely emigrants from the responding LN, for they have completed at least 4 divisions. If they had been activated in the distant LN, some OTII cells that had divided fewer than 4 times would have been observed there from 48h and onwards and these were not present. The primed immigrant OTII cells were present in distant nodes in increased numbers at 96h. These early recirculating OTII cells have features of central memory cells, including expression of CD44 and CD62L. In addition, they re-entered cell cycle and migrated to B cell follicles, much more rapidly than naïve CD4 T cells and could still be induced to produce IL-4 (3).
Conclusions
Analysis of the location of the responding CD4 T cells reveals that while some colonize the B cell follicles, others remain in the outer T zone and yet others leave the draining node and become recirculating cells with central memory-like-features.
In addition, the population of CD4 T cells remaining in the responding LN contains Th2-features as well as TFh-features, and to a lesser extent some properties associated with Th1 cells. The extensive heterogeneity of cytokine producing cells is further exemplified by the finding that some Th2 cells produce both IL-4 and IL-13, while others produce IL-4 but not IL-13, or IL-13 but not IL-4.
The extent of this heterogeneity induced in responses in vivo is only now starting to be appreciated and there is a need to extend studies documenting this diversity and its functional significance.
This information from in vivo model systems is essential to get insight into these molecular events responsible for T cell diversification. Characterising the CD4 T cell diversification occurring during primary immune responses may give clues as to how these cells acquire specific effector functions. This research offers the potential to give clues onto how to regulate the effectiveness of protective immunity and moderate unwanted effects such as autoimmunity and allergy. This may also open up novel ways of designing new forms of vaccines.
References:
1. Serre, K., E. Mohr, F. Gaspal, P. J. Lane, R. Bird, A. F. Cunningham, and I. C. MacLennan. 2010. IL-4 directs both CD4 and CD8 T cells to produce Th2 cytokines in vitro, but only CD4 T cells produce these cytokines in response to alum-precipitated protein in vivo. Mol Immunol 47:1914-1922.
2. Serre, K., E. Mohr, K. M. Toellner, A. F. Cunningham, S. Granjeaud, R. Bird, and I. C. MacLennan. 2008. Molecular differences between the divergent responses of ovalbumin-specific CD4 T cells to alum-precipitated ovalbumin compared to ovalbumin expressed by Salmonella. Mol Immunol 45:3558-3566.
3. Serre, K., E. Mohr, K. M. Toellner, A. F. Cunningham, R. Bird, M. Khan, and I. C. Maclennan. 2009. Early simultaneous production of intranodal CD4 Th2 effectors and recirculating rapidly responding central-memory-like CD4 T cells. Eur J Immunol 39:1573–1586.
4. Smith, K. M., J. M. Brewer, C. M. Rush, J. Riley, and P. Garside. 2004. In vivo generated Th1 cells can migrate to B cell follicles to support B cell responses. J Immunol 173:1640-1646.
5. Peixoto, A., C. Evaristo, I. Munitic, M. Monteiro, A. Charbit, B. Rocha, and H. Veiga-Fernandes. 2007. CD8 single-cell gene coexpression reveals three different effector types present at distinct phases of the immune response. J Exp Med 204:1193-1205.
6. Peixoto, A., M. Monteiro, B. Rocha, and H. Veiga-Fernandes. 2004. Quantification of multiple gene expression in individual cells. Genome Res 14:1938-1947.
7. MacLennan, I. C., A. Gulbranson-Judge, K. M. Toellner, M. Casamayor-Palleja, E. Chan, D. M. Sze, S. A. Luther, and H. A. Orbea. 1997. The changing preference of T and B cells for partners as T-dependent antibody responses develop. Immunol Rev 156:53-66.
8. Garside, P., E. Ingulli, R. R. Merica, J. G. Johnson, R. J. Noelle, and M. K. Jenkins. 1998. Visualization of specific B and T lymphocyte interactions in the lymph node. Science 281:96-99.